Our lab has generated a highly versatile functional imaging platform that utilizes DNA nanotechnology to chemically map sub-cellular structures known as organelles. We seek to answer fundamental questions such as how the chemical composition of an organelle, which has been optimized over evolutionary timescales, impacts its function at the sub-cellular level. How do these changes alter cell function, tissue function, and organism physiology? We utilize bionanotechnology to investigate these questions by exploiting our understanding of cellular machinery to develop solutions to problems in science and engineering. Our research takes advantage of nucleic acid structure and dynamics to create DNA-based nanodevices for quantitative chemical imaging of living systems.
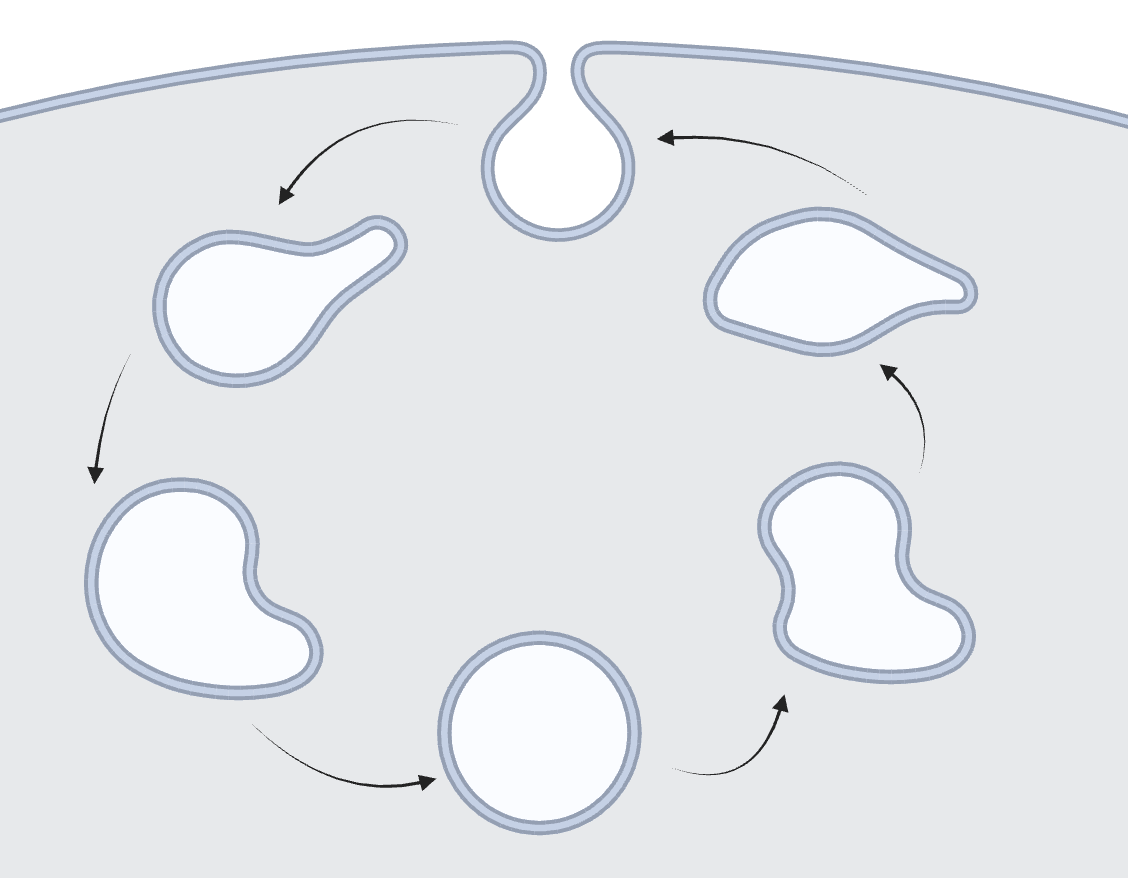 |
Mapping pH of the Endosomal Pathway The endosomal pathway contains early endosomes, late endosomes, and lysosomes. It is a highly dynamic environment used for transporting cargo into various cellular compartments. With the creation of the I-switch, our lab measured the pH of various compartments along the endosomal pathway to further understand how pH is related to the maturation of endosomes within the cell. Sodium and potassium measurements using our nanodevices RatiNa and pHlicKer, respectively, have further supported our understanding of endosomal maturation by mapping the ion levels of endosomes as they mature. |
|||
|
Focusing on Lysosomes and Recycling Endosomes Lysosomes are a key player in the endosomal pathway and are responsible for the breakdown of biological material within cellular environments. Our work has led to the characterization of lysosomal lumenal pH and chloride, as well as reactive species such as hypochlorous acid (HOCL). Recently, we have also uncovered the calcium levels of this organelle, as well as elucidating a mechanism for calcium entry into the lysosome with our nanodevice, CalipHlour. In the recycling endosome, another step along the endosomal pathway, we have used our DNA nanodevices to quantify parameters such as pH, chloride, and membrane potential. |
 |
|||
 |
The Trans-Golgi Network The trans-Golgi network (TGN) contributes to the sorting, modifying, and tagging of proteins from the endoplasmic reticulum and is part of the retrograde pathway. We once again deployed the I-switch to measure pH in the TGN. We have also used another nanodevice, NOckout, to measure NO levels and uncover the mechanisms of Nitric oxide synthase 3 (NOS3). Recently, with the creation of pHlicKer, we have measured potassium levels within the TGN in a pH-independent manner. |
|||
|
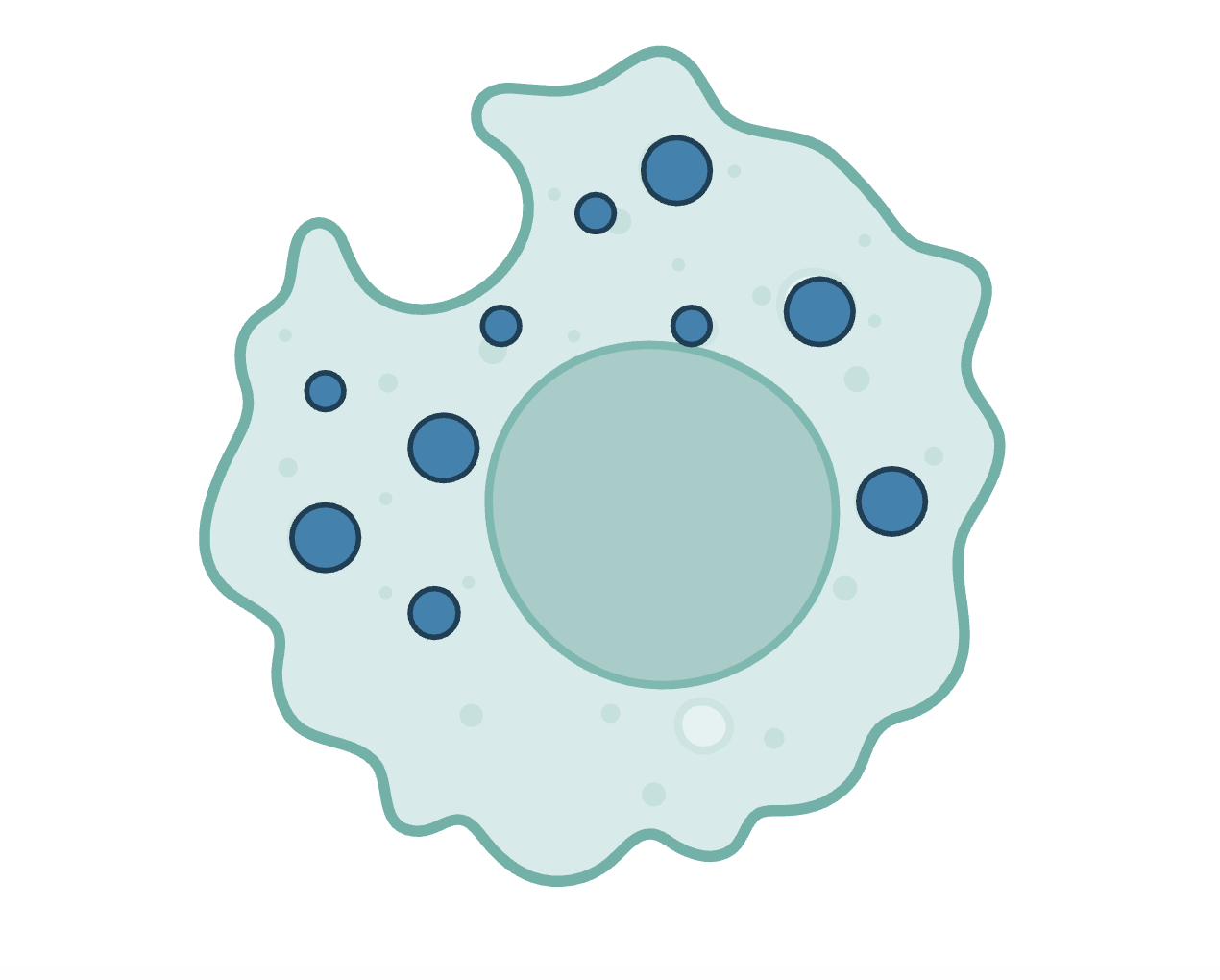
Phagosomes Phagosomes are the product of immune cell engulfment of foreign debris. We created a DNA nanodevice, z-cHOClate, to measure pH and HOCl levels in the phagosomes of macrophages, monocytes, and neutrophils. We have also started work in other areas of immunology, such as using DNA nanodevices to deliver cathepsin inhibitors to the lysosomes of tumor-associated macrophages (TAMs). |
 |
|||
 |
Mapping Organelles in vitro With the use of C. elegans and zebrafish, we have been able to target our devices in a tissue-specific manner. For example, the development of new synthetic receptors in C. elegans has allowed us to specifically target neuronal plasma membranes, endolysosomes of cholinergic or glutamatergic neurons, or lysosomes of intestinal epithelial cells. We also developed two new nanodevices for C. elegans chloride ion measurement: Clensor, which is pH-insensitive, and ChloropHore, which maps chloride and pH simultaneously. With the use of NOckout in zebrafish, we were also able to quantify NOS2 activity in the phagosomes of zebrafish microglia. |